Input file reference manual¶
DockOnSurf execution is controlled by the edition of an input file. In this
file, all the mandatory and optional parameters are specified using a keyword
= value pair. An example of such file is provided in
dockonsurf/examples/dockonsurf.inp.
Keywords must be specified under one of the four existing sections: Global, Isolated, Screening and Refinement , the last three corresponding to the stages DockOnSurf consist on. Keywords can beclassified in three categories:
mandatory, for which its value must always be provided (eg. run_type)
keyword-dependent, for which specification is only mandatory depending on the value of another keyword (eg. subm_script keyword is mandatory if batch_q_sys has a value different than “False”)
optional, for which a default value is assigned when the keyword is not given any value (eg. max_structures).
Although none of the three stages is compulsory for a DockOnSurf execution, the necessary keywords for each of the stages will be also considered as mandatory. They are mandatory in the case of running such stage.
For some keywords, the use of more than one value is possible. In order to separate such values, a whitespace (” “), a comma (“,”) or a semicolon (“;”) can all be used. Some values need to be specified in groups (eg. each of the three vectors of the cell matrix). In such case groups are defined by enclosing the elements around parentheses-like characters: “()”, “[]” or “{}”. (eg. “(a1, a2,a3) [b1 b2 b3] {c1;c2; c3}”).
Global¶
Parameters in this section have an effect on the entire DockOnSurf execution.
Note
When using VASP as code, the PBC cell must be provided either through the pbc_cell keyword or inside the coordinates file. (eg. POSCAR/CONTCAR)
Isolated¶
Parameters in this section have an effect on the Isolated stage.
Screening¶
Parameters in this section have an effect on the Screening stage.
Warning
If both, the collision_threshold and min_coll_height are set to “False”, atomic clashes will NOT be checked.
Warning
When max_structures is set, structures are chosen randomly and reproducibility is not kept.
Warning
If both the collision_threshold and min_coll_height are set to “False”, atomic clashes will NOT be checked.
Note
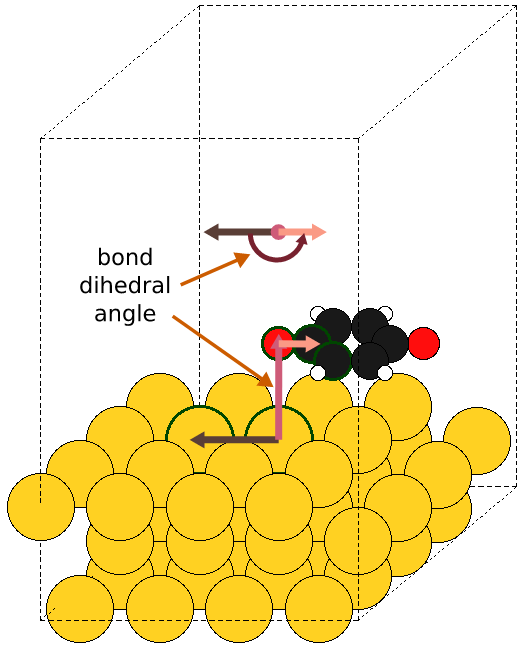
All possible combinations of the three different angles are generated, leading to \(n^3\) raw configurations. However, for both, the Euler (in its x-convention) and the internal set of angles, redundant configurations are generated with a dependence on the rotations per angle \(n\)) equal to \(n(n-1)\): In the x-convention of Euler angles, \((x, 0, 0) \equiv (0, 0 , x)\). In the internal angles, when the bond angle is flat, all the bond-dihedral angle rotations are ineffective. These redundant configurations generated both in the Euler and Internal angles are algorithmically filtered out.

Refinement¶
Parameters in this section have an effect on the Refinement stage.
Note
The energy_cutoff must be chosen considering the approximate adsorption energy. For a given value several low-interacting species may fall within the energy cutoff, while for the most binding ones, only few of them may be selected for refinement.
- Versions
- latest
- Downloads
- On Read the Docs
- Project Home
- Builds
Free document hosting provided by Read the Docs.